Research...
“There is nothing like looking, if you want to find something...
You certainly usually find something, if you look, but it is not always quite the something you were after.”
“Simplicity does not precede complexity, but follows it.”
On this page, I will discuss my primary research, which brings together ideas from intelligent search and complexity science. Specifically, it involves the use of evolutionary search processes to explore multi-agent simulations of complex adaptive systems.
Page Outline:
- Background
- Query-based model exploration
- Case study: finding flocking
- Case study: viral marketing strategies
- Calibration & sensitivity analysis
- Sampling noisy fitness landscapes
- CrossNet: network-based chromosomal structures
The software design component (BehaviorSearch) of my thesis is discussed on the tools page, and various other (non-thesis) projects are on the projects page.

Ray-traced visualization of a viral marketing simulation on part of the Twitter network. (full size)
Featured as cover art for the ASME's Mechanical Engineering Magazine, March 2012!
Background
What are “complex systems”?
Complex systems pose a challenge to the long-standing reductionist approach to science. A complex system is composed of multiple interacting units, but if you take it apart and study the units individually, this gives little insight into the behavior of the system as a whole. Even in cases where the individual units follow very simple rules, complex patterns of behavior can emerge from the interactions between the individuals. When the whole system's behavior appears to be “more than the sum of its parts”, this is called emergence. For example, the efficient foraging capability of an ant colony is an emergent behavior, based on individual ants depositing and interacting with each other via pheromone trails (see video at right).
Complex systems researchers study commonalities of emergent interaction patterns across many disciplines. A few examples of complex systems include the brain (composed of interacting neurons), economic markets (composed of interacting firms), the U.S. power grid, food webs, metabolic pathways, disease epidemics, and the evolution of language.
What are “multi-agent simulations”?
Multi-agent simulation (a.k.a. agent-based modeling or individual-based modeling) is one of the foremost techniques for investigating complex systems. In this sub-genre of computer simulation, each virtual agent is given a set of rules, and then multiple agents interact according to those rules.
The Defend and Flee video at right shows an example of how a group of agents following simple rules can generate a a variety of complex behaviors. Each agent perceives one other agent as a friend, and another as an enemy (both randomly chosen). Red agents try to move between their friend and enemy (to defend their friend), while blue agents try to move behind their friend (to flee from their enemy). As the video shows, simulations starting with different random initial conditions can result in qualitatively different behaviors.
What is “evolutionary computation”?
The field of evolutionary computation consists of a family of nature-inspired search/problem solving techniques tracing back to the 1950s. Evolutionary algorithms seek to harness the power of Darwinian evolution to find solutions to challenging search/optimization problems on the computer. I believe evolutionary algorithms are well-suited for exploring problems in multi-agent systems, where the complex interactions and feedback processes can lead to challenging search spaces. Through the coupled processes of variation and selection, genetic algorithms sometimes evolve clever solutions that human researchers would otherwise miss.
Simulated ant foraging (red ants, green/white pheromones).
For more info, see the Ants model in the NetLogo models library.
An agent-based model showing how complex behavior can emerge from simple rules.
Simple Genetic Algorithm demo that I wrote, available in the NetLogo models library.
Thesis Research
Query-based model exploration
When researchers develop agent-based models, they define a potentially large number of parameters that control aspects of the agents or their environment (e.g., ant metabolism rate, wind speed, pheromone dissipation rate, etc.). To fully understand the model, the impact of these parameters must be understood, and the range of possible outcomes must be explored.
Each parameter can affect the system's behavior in different ways, and the interactions between multiple parameters can cause unexpected (and nonlinear) results. Supposing that there are 10 parameters, 10 choices for setting each parameter, and each simulation run takes 10 seconds, then exhaustively exploring the space of possible parameters would take more than 3000 years. (Furthermore, the model must be run multiple times for each parameter setting combination to determine the range of behavior possible, since agent-based models almost always have stochastic elements or initial conditions.)
Running the simulation answers the question “What is the system behavior for a given set of parameters? ”. My primary research focuses on efficient methods of answering the inverse question: “How can I find parameter settings that will cause a specific system-level behavior to occur? ”. I refer to this approach as query-based model exploration, and my primary thesis work applies evolutionary search techniques to address this problem.

Nonlinear parameter interactions in a predator prey model.
(Although much variation stems from model stochasticity, the underlying trends are also nonlinear.)
Case study: finding flocking
As an example domain, I consider agent-based models of the dynamics of collective animal movement (i.e. flocking, schooling, swarming). Most work in this area traces back to Reynolds' seminal work on developing realistic-looking flocking behavior for computer-generated/cinematic animation, which he playfully dubbed boids.
To see natural flocking in action, an Internet search will quickly turn up examples like this clip of starlings in flight near Otmoor wetlands in the U.K.:
Internet video of flocking starlings
NetLogo's model library includes a variant of the boids model, simply called Flocking. There is also a recent addition to the models library called Flocking Vee Formations, which I helped my colleague Michelle Wilkerson-Jerde to develop, based on ideas from Nathan & Barbosa.
In a case study using these models, I demonstrated the use of several search algorithms (including genetic algorithms) on four different exploration tasks: searching for convergence, non-convergence, volatility, and the ability to form vee/eschelon shapes (such as those observed in Canada geese). Further details of this study may be found in this paper.
In the NetLogo Flocking model, birds can bend and weave their way into groups.

Photo: flying geese

Measuring simulated patterns in the Flocking Vee Formation agent-based model
Case study: viral marketing strategies
In joint work with Bill Rand and my advisor Uri Wilensky, I used genetic algorithms to search the parameter-space of an agent-based diffusion-of-innovation model, in order to discover good seeding strategies for viral marketing. We examined five social network structures (see image at right): four abstract and artificially generated, and one empirically extracted from Twitter friendship/following data.
The following (artistically ray-traced) visualization video of the agent-based model portrays how the adoption of a product might cascade through a 1000-node Twitter subgraph. Agents that haven't adopted the product yet are shown as birds on white spheres, and agents that have adopted the new product are shown as hat-wearing birds on purple spheres.
This work uncovered interesting differences between seeding strategies for the empirical Twitter-based network, relative to the abstract network models. Full discussion and results can be found in this paper.

Network visualizations: Erdős–Rényi random graph, lattice, preferential attachment, small-world, Twitter subnetwork
Calibration & sensitivity analysis
I also demonstrated the use of genetic algorithms for several important model analysis tasks in the context of the well-known Artificial Anasazi agent-based model, which uses simulation to explore an archaeological mystery about why the Anasazi people abandoned a region of settlement after hundreds of years of occupation. (For a NetLogo-based replication of this model, see openabm.org.)
In this work, which is particularly inspired by John Miller's work on active nonlinear testing, I showed how genetic algorithms could calibrate the model parameters to best match archaeological data, and also how genetic algorithms could perform sensitivity analysis by searching for nearby parameters that yield a poor fit with the data. In addition, this approach to sensitivity analysis led us to discover a small bug in the model's code.
To find out more, see this paper.
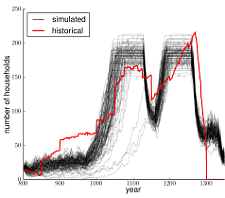
Application: Evolving simulated river topographies
I've also applied similar techniques in collaboration with my wife, Dr. Susa Stonedahl, to evolve the parameters (technically Fourier coefficients) that describe a river's topography in order to best match a certain hyporheic profile, characterized by how long particles spend traveling between the surface water and the groundwater underneath the stream. This work offers insight into the complex mapping between topographies & hyporheic flow paths.
To find out more, read our Darwinian Rivers paper.
Sampling noisy fitness landscapes
One of the specific challenges of using genetic algorithms to search parameter-spaces is that agent-based models almost always incorporate random elements. The initial position and/or orientation of agents is often chosen randomly, the scheduling of agent actions is typically in randomized order, and agents commonly use probabilistic rules. Thus, running the agent-based model multiple times will yield differing results, even with the exact same model parameters. When using genetic algorithms to search for a certain behavior, this effectively means that the fitness function is noisy.
Repeated sampling can be used to reduce noise, but how much sampling is worthwhile? The problem is compounded when using a technique called “fitness caching“, which saves the results from previous model runs so they do not have to be re-computed; caching causes the noise in the landscape to become frozen. In this paper, I discuss the interaction between noise and fitness caching, and derive several heuristics that may be useful in choosing an appropriate amount of sampling for noise reduction.
Noise can cause a smooth fitness landscape to exhibit many jagged peaks, which may confound search algorithms that are looking for the true peak.
CrossNet: network-based chromosomal structures
Another important challenge for efficient evolutionary search is the incorporation of domain-specific knowledge into the search process. Specifically, human researchers often develop many intuitions about how the parameters of an agent-based model affect its behavior. While it is possible that some of these intuitions may be wrong, many of them will be correct, and it could be beneficial to use this information to increase the efficiency and effectiveness of an automated search process such as genetic algorithms.
One potential way to include a priori knowledge or intuitions about the search space into the genetic algorithms is by modifying the chromosomal structure. Traditional genetic algorithms use a linear structure for storing genotypic information, and genetic crossover (recombination) operates on two such linear structures. In this paper, I propose the CrossNet recombination framework, which uses a network to represent linkage between genes, and test it out on hyperplane defined functions and evolving rules for cellular automata.

A simple contagion model on the network chooses a mask to determine which genes get swapped during crossover.
Quick Links: